Restriction Enzymes: A Brief History
Restriction enzymes, or restriction endonucleases, are proteins that recognize and cleave DNA at or around specific sites.
The discovery of restriction enzymes originates from the ongoing battle for life between bacteria and bacteriophages.
Bacteriophages are small viruses that inject DNA into a host bacterium. Once the viral DNA is inside the host, it utilizes the replication machinery to create thousands of bacteriophage progeny. In many cases, the resulting viral replication inside the host causes its lytic destruction.
In the 1960s, Werner Arber observed how bacteria evolved a restriction and modification system to protect themselves from bacteriophage infection. Restriction involves the breakdown of DNA, whereas modification describes a chemical process (DNA methylation) that protects DNA from restriction.
Hamilton O Smith later isolated the enzymes involved in the restriction and modification system, which ultimately confirmed Arber’s hypothesis. Smith’s pioneering work was published in 1970 and is the first description of a restriction enzyme, which was isolated from the bacterium Haemophilus influenzae.
With the discovery of restriction enzymes, Smith’s colleague Daniel Nathans explored the applications of these proteins to genetics. In 1971, Nathans described how specific restriction enzymes can cleave larger pieces of DNA, SV40 in this case, into smaller fragments. Two years later, the team demonstrated the utility of restriction enzymes to create a cleavage map of a viral genome.
In 1978, Werner Arber, Daniel Nathans and Hamilton O Smith were jointly awarded The Nobel Prize in Physiology or Medicine for their discovery of restriction enzymes and their application to the problems of molecular genetics.
Restriction Enzyme Nomenclature
According to The Restriction Enzyme Database (REBASE), there are well over 5,000 known restriction enzymes!
Restriction enzymes are named after the source microorganism that they are extracted from. Specifically, the name includes the genus, species, strain abbreviation and order of discovery in that strain. The order is denoted as a Roman numeral.
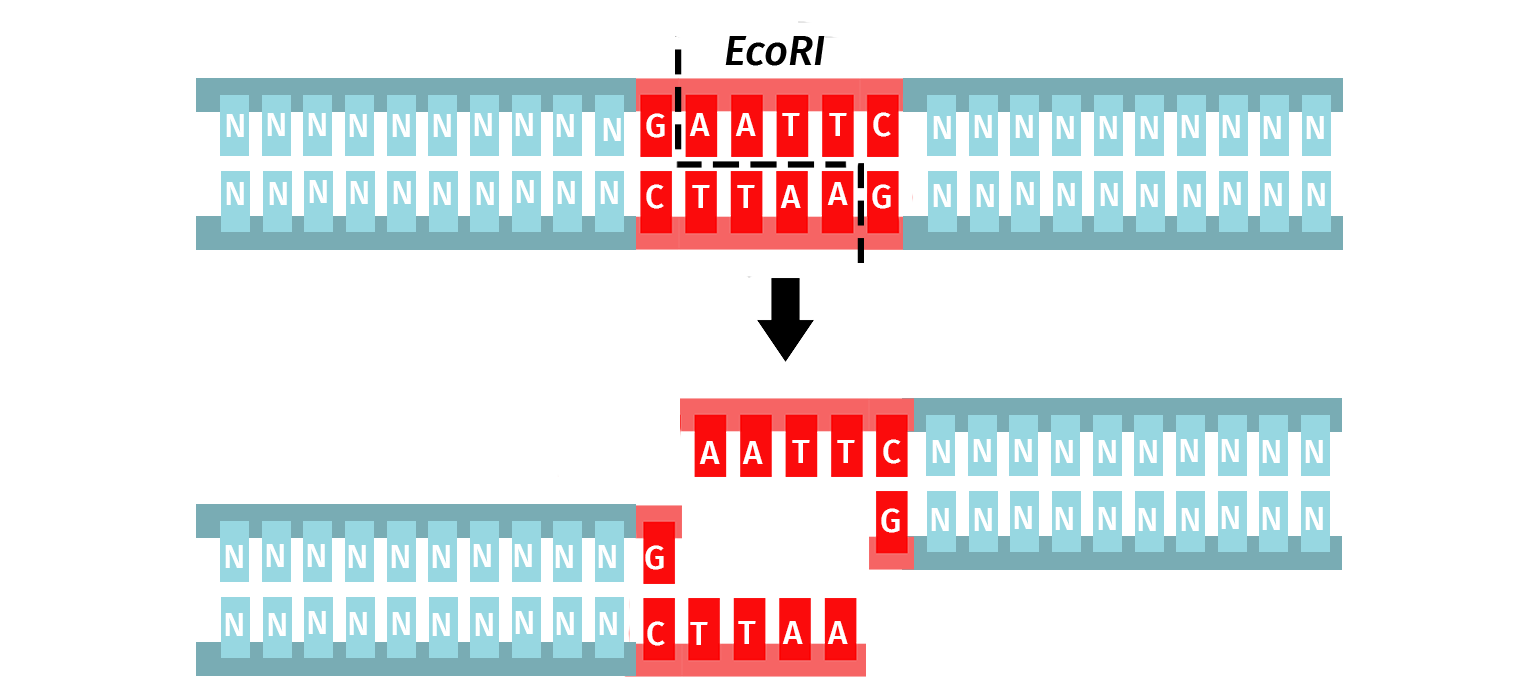
For example, the enzyme EcoRI is used since it has the following description:
- Genus: Escherichia
- Species: coli
- Strain: R (denotes the strain RY13)
- Order: I (first restriction enzyme discovered in the strain)
The table below provides the nomenclature of some common restriction enzymes.

Restriction Enzyme Types
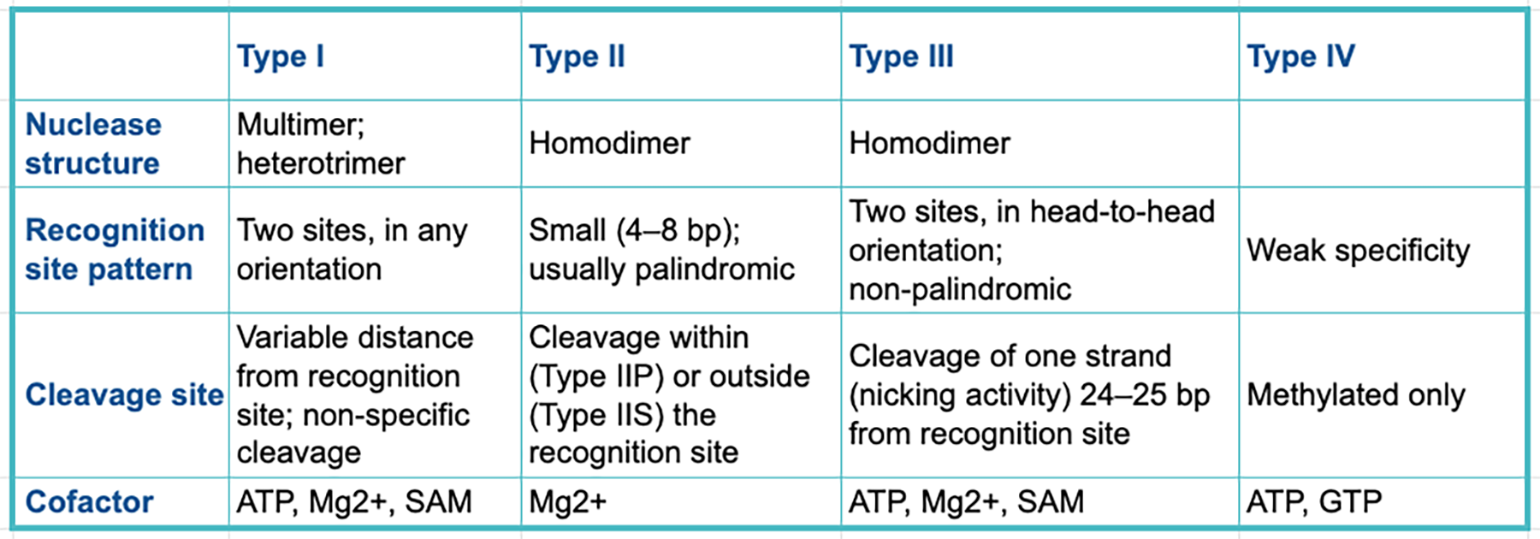
Restriction enzymes can be categorized into four classes known as types. Each type has a distinct structure, recognition site pattern, cleavage approach and desired cofactors.
The table below summarizes the main differences between the key restriction enzyme types.
Type I Restriction Enzymes
Type I enzymes are multifunctional proteins with three separate subunits that are involved in restriction, modification (methylation) and specificity (DNA recognition).
Examples of Type I restriction enzymes include EcoAI, EcoKI and StySBLI.
Type II Restriction Enzymes
Type II enzymes are the most abundant and well-characterized of all restriction enzyme categories.
Within the Type II category, there are up to 8 different subdivisions that have slightly different recognition site and cleavage properties. Type IIP and Type IIS are the most common subdivisions associated with molecular biology applications.
Type IIP Restriction Enzymes
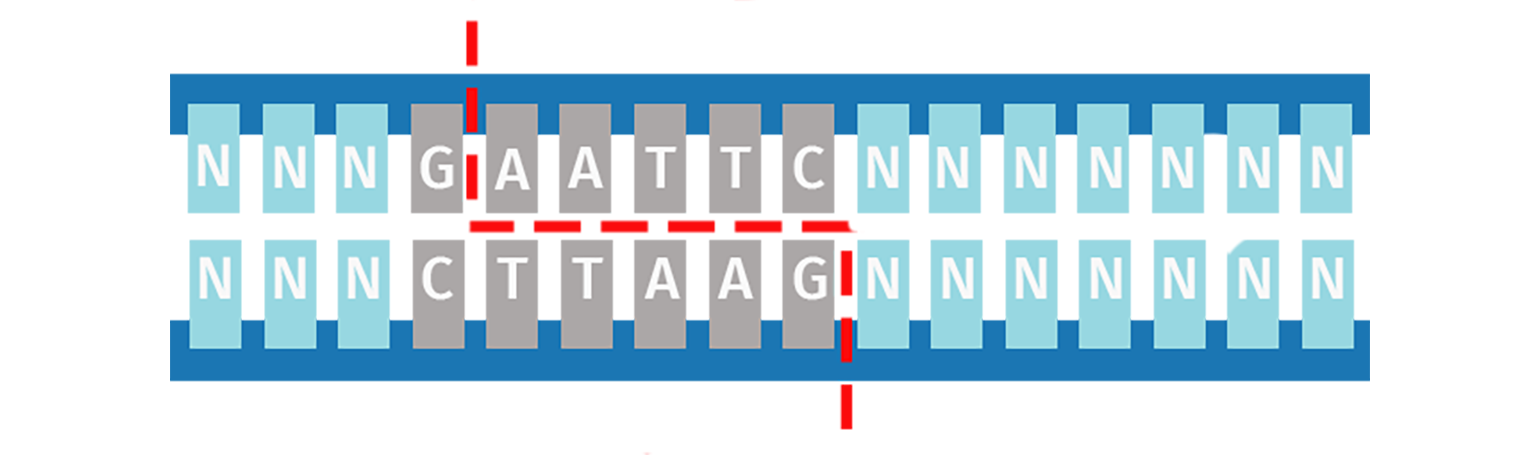
The orthodox Type IIP enzymes recognize a palindromic sequence (4–8 bp) and cleave both DNA strands within or immediately adjacent to the recognition site. The result is a product with either sticky (5′ or 3′ overhangs) or blunt ends.
The versatility of Type IIP restriction enzymes makes them useful for conventional restriction cloning, where a vector and an insert are digested with the same enzyme pairs to create compatible ends that can be ligated together.
► Learn more about restriction enzyme cloning
Type IIS Restriction Enzymes
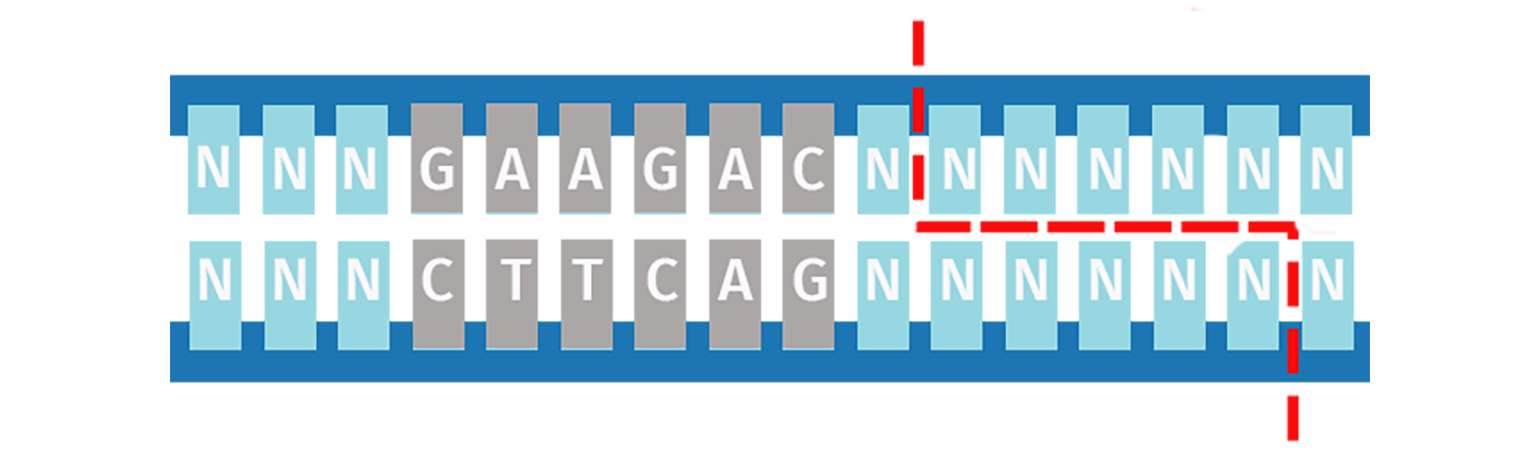
Unlike Type IIP restriction enzymes, the Type IIS subdivision recognizes a non-palindromic sequence and cuts DNA at a defined distance downstream of the recognition site. Cleavage is staggered on each strand and is sequence-independent, which means the same Type IIS enzyme can produce different sticky ends.
Type IIS restriction enzymes are utilized in Golden Gate Assembly to create the ordered assembly of one or more DNA fragments in a single-step cloning procedure.
► Learn more about Golden Gate Assembly
Type III Restriction Enzymes
Type III restriction enzymes contain a restriction and modification complex that acts as a single protein unit. They require two recognition sequences (5–6 bp) that are non-palindromic for successful cleavage.
There is only a single Type III restriction enzyme - EcoP15I - that is commercially available.
Type IV Restriction Enzymes
Type IV restriction enzymes only cleave methylated DNA and will not act upon unmethylated DNA.
EcoKMcrBC, the only Type IV restriction enzyme commercially available, requires two half-sites for successful DNA recognition; these sites can be separated by up to 3 kb!
Restriction Enzymes and DNA Methylation
Some restriction enzymes are sensitive to DNA methylation modifications. Specifically, when methylation occurs within the recognition site, it can directly prevent the enzyme from cleaving DNA.
It’s important to appreciate the source of DNA during cloning experiments since the location and amount of DNA methylation varies between organisms.
DNA Methylation in Eukaryotes
Some eukaryotes, such as mammals, utilize DNA methyltransferases to manage their chromatin and control gene expression.
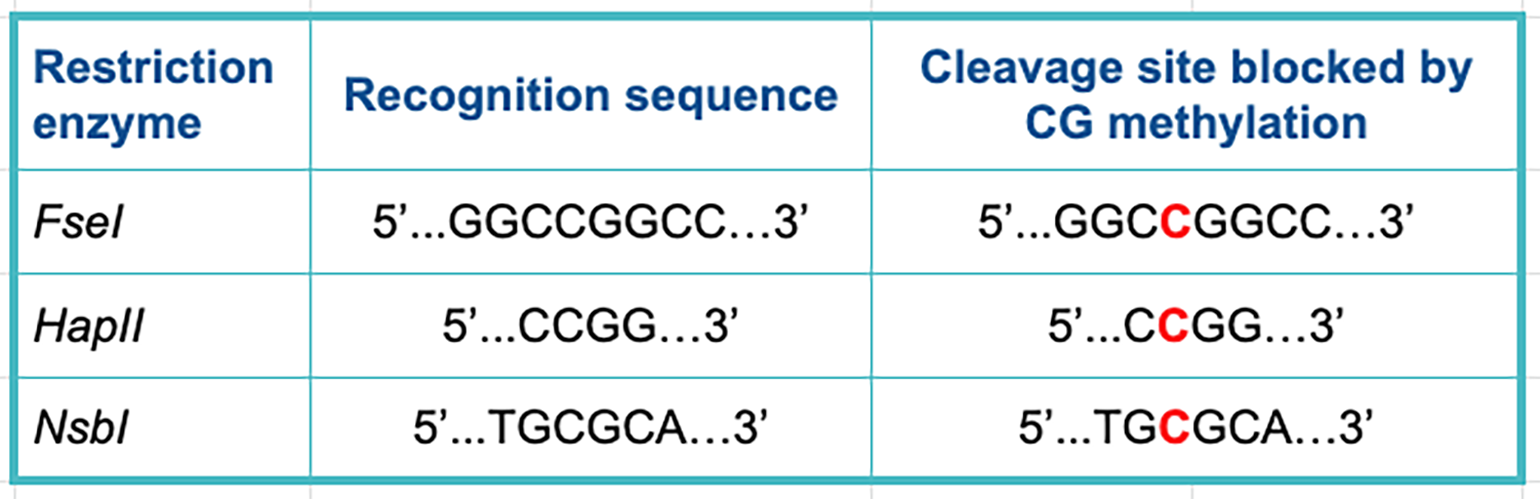
The most common location of DNA methylation in mammals occurs at CpG sites, whereby the cytosine is methylated when adjacent to a guanine.
The table below lists some examples of restriction enzymes that can be affected by CG methyltransferase enzymes. Methylated bases in the cleavage site are marked in red.
DNA Methylation in E. coli
Most strains of E. coli have three site-specific DNA methyltransferases: Dam, Dcm and EcoKI.
- Dam: Methylates the adenine base in the sequence 5’...GATC…3’
- Dcm: Methylates the internal cytosine base in the sequence 5’...CCWGG…3’ (W refers to either an A or T base)
- EcoKI: Methylates the adenine base in the sequence 5’...AACNNNNNNGTGC…3’ and 5’...GCACNNNNNNGTT…3’ (N refers to any base)
DNA Methylation Following in vitro DNA Synthesis
In vitro synthesis of DNA, such as via PCR, Gibson Assembly, and In-Fusion® Cloning, are performed in closed reactions in the absence of DNA methyltransferases, therefore the resulting products will contain no methylation.
DpnI and DNA Methylation
Unlike most other restriction enzymes, DpnI is only active when its recognition sequence is methylated. Specifically, DpnI requires the adenine base to be methylated in the sequence 5’...GATC…3’.
The unique properties of DpnI make it a useful tool to selectively eliminate methylated template DNA. For example, DpnI can be used to digest unwanted plasmid DNA originating from Dam+ E.coli that is left over following site-directed mutagenesis.
Learn More About Cloning