How to сlone with Topoisomerase I?
TOPO cloning is a one-step molecular cloning method used to insert DNA fragments into linearized vectors via the actions of a single enzyme - topoisomerase I.
In molecular biology, Topoisomerase I, extracted from the Vaccinia virus, functions as both a restriction enzyme and a DNA ligase to enable cleavage and rejoining of DNA during replication.
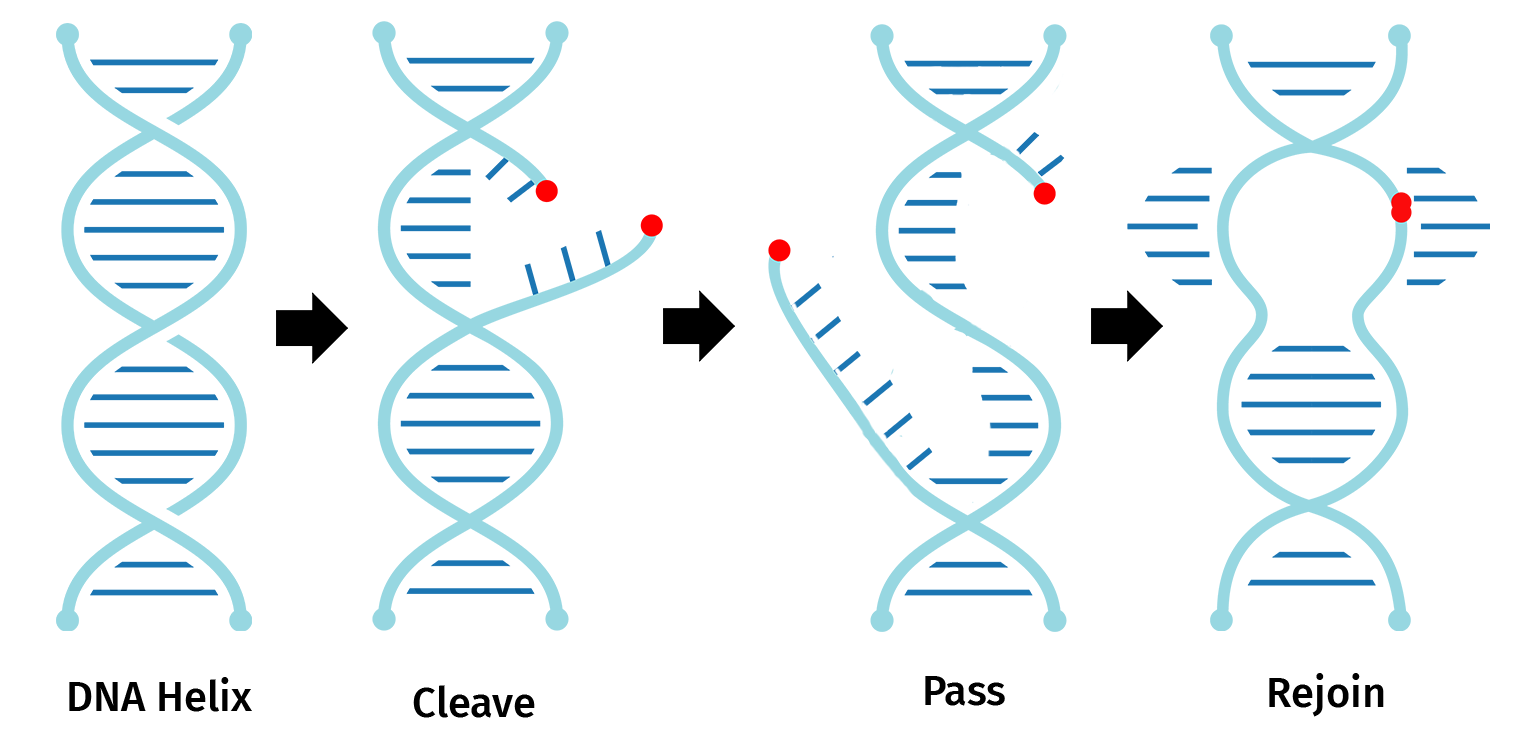
The restriction domain of topoisomerase I recognizes and cleaves a single DNA strand at the pentameric sequence 5’...(C/T)CCTT…3’. The enzyme then forms a covalent bond with the phosphate at the 3’ thymidine.
Topoisomerase I bound to the 3’ phosphate group of thymidine is also able to religate the nicked DNA.
The unique properties of topoisomerase I have been exploited to form the basis of TOPO® cloning.
Types of Cloning with Topoisomerase I
There are three types of Topoisomerase I:
- TA-TOPO cloning
- Blunt-end TOPO cloning
- Directional TOPO cloning
Both TA and Blunt-end cloning are bidirectional with inserts able to ligate in either orientation into the vector. Directional TOPO cloning, as the name suggests, ensures inserts ligate in a preferred orientation.
TA or Sticky-End Cloning
TA, also known as sticky-end, TOPO cloning exploits the inherent activity of Taq polymerase, which adds a single deoxyadenosine (A) to the 3'-end of PCR products.
When inserts are amplified by PCR with Taq polymerase, the product can be ligated into suitable TA TOPO cloning vectors. These vectors are prepared in advance so that they come linearized and with the topoisomerase I enzyme covalently bound to the free 3’ thymidine overhangs.
The insert and vector are simply mixed together to enable the overhangs to ligate. The ligation with topoisomerase I is performed in as little as 5 minutes at room temperature; this is in contrast to T4 DNA ligase that is typically used in restriction cloning, which requires incubation on ice for 30–60 minutes for successful ligation.
► Learn more about restriction cloning
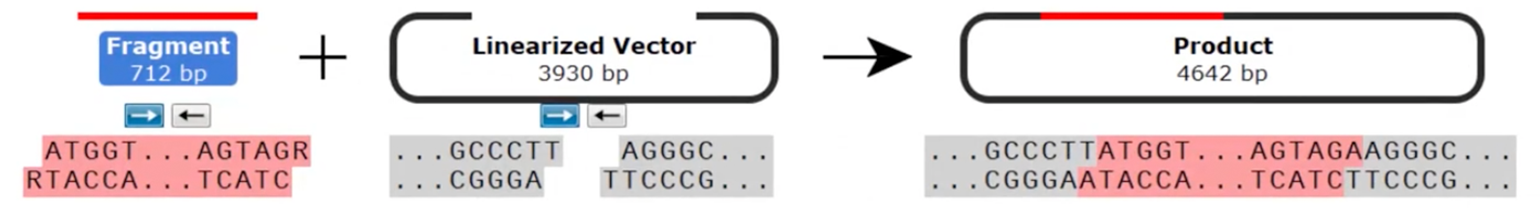
Despite the single base overhang, TA TOPO cloning is not considered directional since the insert can be ligated into the vector in either orientation.
► Learn to simulate TA TOPO cloning in SnapGene.
Blunt-End Cloning
In Blunt TOPO cloning, the insert is commonly created through PCR with high-fidelity polymerases that contain 3’ to 5’ exonuclease activity, thus leaving blunt-ended products. Alternatively, inserts are digested with restriction enzymes that form a blunt end.
Commercial Blunt TOPO cloning vectors come linearized with blunt ends that contain the sequence 5’...CCCTT 3’.
Mixing the blunt-ended insert and linearized vector with topoisomerase I enables ligation.
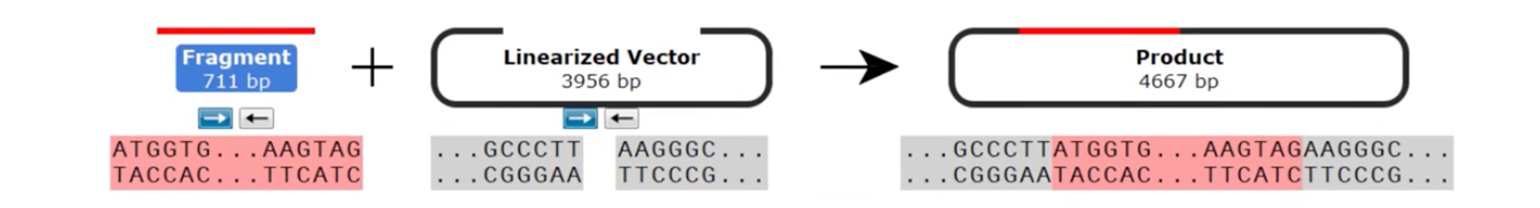
As with TA TOPO cloning, Blunt TOPO cloning is also non-directional due to the presence of blunt ends on both the insert and vector.
► Learn to simulate Blunt TOPO cloning in SnapGene
Directional TOPO Cloning
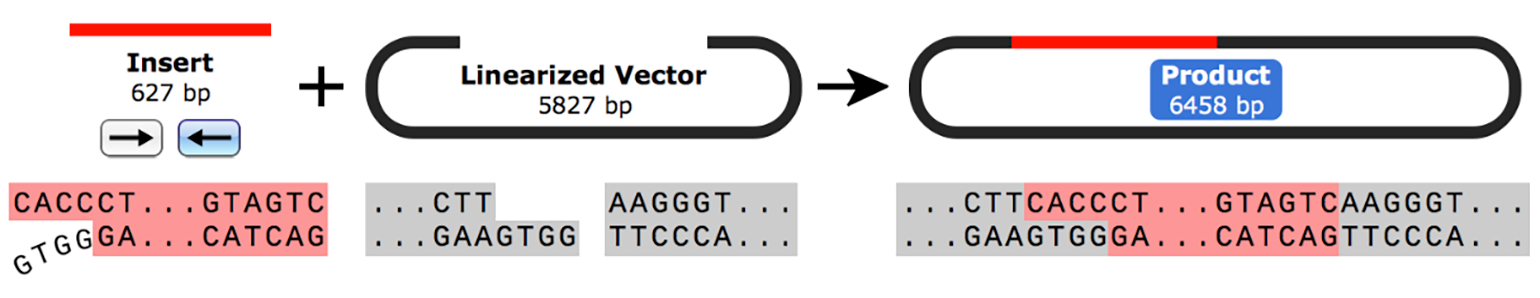
In Directional TOPO cloning, the insert is designed so that it contains a CACC overhang at the 5’ end and a blunt end at the 3’ end. The four-nucleotide 5’ overhang is usually introduced by PCR with a modified primer and is essential for the directional component of the technique.
On the other hand, the Directional TOPO cloning linearized vector contains a 5’ GTGG overhang with a 3’ blunt end. Topoisomerase I ligates the insert and vector in the desired orientation according to the overhang placement.
► Learn to simulate Directional TOPO cloning in SnapGene
Tips and Additional Resources
Primers Should Not Contain 5’ Phosphates
When designing primers for TOPO cloning, ensure that there are no phosphates added to the 5’ end. Topoisomerase I requires the free hydroxyl group for successful ligation.
Allow Sufficient PCR Extension Time Prior to TA TOPO Cloning
Since TA TOPO cloning relies on the presence of a single 3’ A overhang on the PCR insert, you need to ensure the final extension step of the PCR allows sufficient time for Taq polymerase to incorporate the overhang into the insert.
Consider Adjusting the Topoisomerase I Incubation Time
For most TOPO cloning experiments, a final 5-minute incubation at room temperature is sufficient to allow topoisomerase I to successfully ligate the insert and vector. You may want to consider extending the incubation time to 20–30 minutes when using dilute amounts of insert or when the insert is large in size.
If using longer incubation times, be sure to also include salt (e.g. 200 mM NaCl and 10 mM MgCl2) in the reaction buffer. The addition of salt prevents topoisomerase I from rebinding to and cleaving DNA after the ligation step has finished.
Choose the Correct DNA Polymerase for Your Experiment
The type of DNA polymerase will generally depend on your experimental needs. When cloning large fragments, consider using a high-fidelity polymerase with proofreading capabilities to reduce the likelihood of sequence errors.
Remember that proofreading polymerases do not incorporate the 3’ A overhang required for TA TOPO cloning. Therefore, in this instance, you could use a polymerase mixture containing a proofreading enzyme with some conventional Taq to ensure the overhang is still incorporated without compromising the product error rate.

