SnapGene Version 6.2.2
SnapGene 6.2.2 was released on January 09, 2023.
Overview:
Version 6.2 adds support for suboptimal RNA secondary structures and includes a number of other enhancements to the Structure view, includes a number of improvements to the Golden Gate cloning simulation tools and PCR simulations in general, provides better flexibility when working with Protein properties, and modernized buttons and icons.
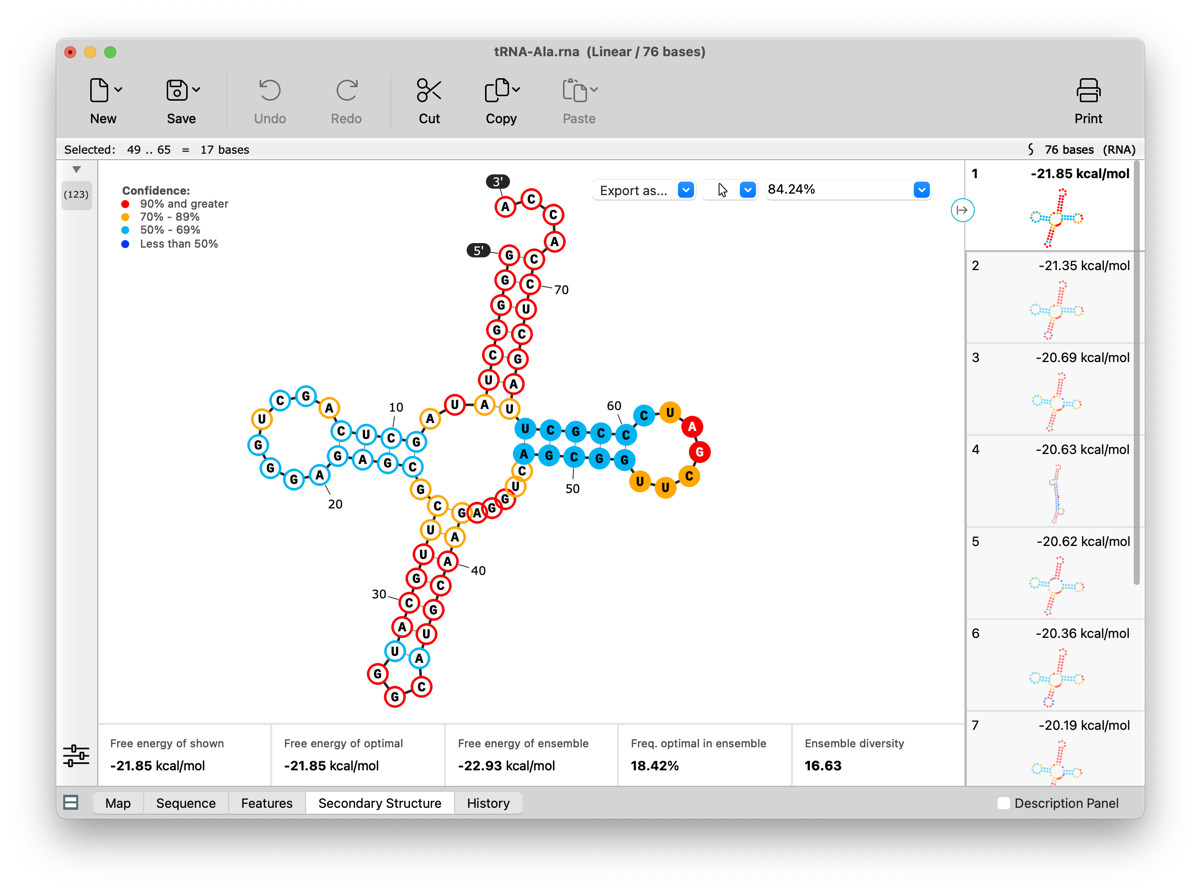
RNA Secondary Structure view enhancements
Suboptimal RNA secondary structures are now shown. All structures can be recalculated using adjusted Tm and other settings. Visualization and selection capabilities have been enhanced, including the ability to display and make sequence selections that are synced across all views. Coordinates and 5' / 3' end labels can optionally be displayed. See View Predicted RNA Secondary Structure for more information.
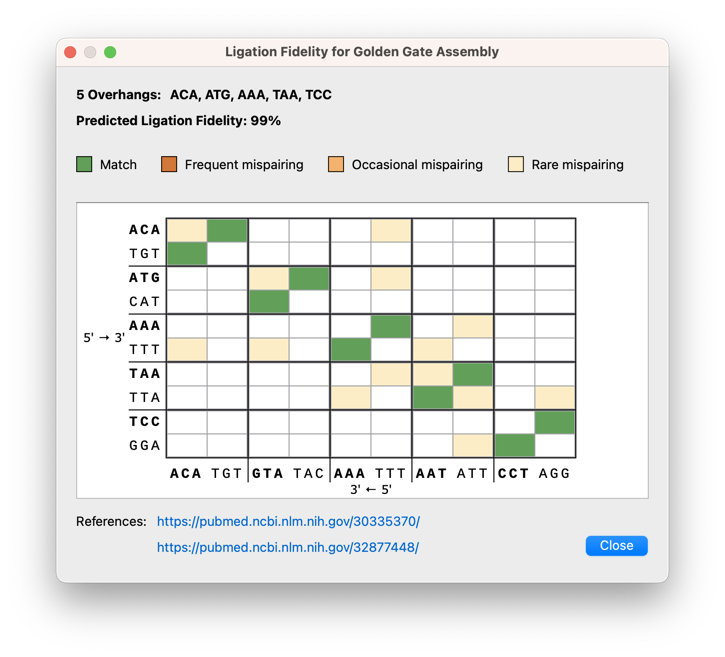
Golden Gate Assembly Improvements
Various enhancements to the Golden Gate cloning dialogs include a ligation fidelity matrix for assessing overall reaction fidelity, it is now possible to adjust overhangs for manually designed primers, a new option to cut the vector with any enzymes (not just Type IIS enzymes), as well as an updated settings dialog with easy access to recommended enzymes.
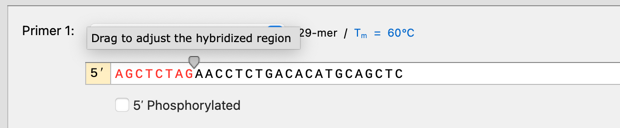
Improvements to primer design in PCR cloning simulations
You can now adjust the hybridization region for all PCR cloning simulations (e.g. PCR, Golden Gate, Gibson, In Fusion, etc.). When using automatic primer design the hybridized region will be configured automatically, ensuring miscellaneous features are not transferred to the product when a 5’ primer extension by chance partially hybridizes to the template.
Protein properties can now be copied and exported
Copy or export the properties and amino acid data for full protein sequences or selected regions in text or spreadsheet (*.csv / *.tsv) formats. Individual properties can also be copied directly from the view.
Modernized toolbar, application, and document icons
SnapGene now uses modernized icons in the top toolbar. The application icon and file icons have also been updated.
New Functionality:
- Suboptimal RNA secondary structures
- RNA structures can be recalculated using adjusted Tm and other settings
- Display and make sequence selections in Structure view
- Coordinates and 5' / 3' end labels can optionally be displayed in Structure view
- Added menu actions at the top right of Structure view to reset pane, rotation and scale
- Added a ligation fidelity matrix to the Golden Gate Assembly tool for assessing overall reaction fidelity
- Cut a Golden Gate Assembly vector with any enzyme (not just Type IIS enzymes)
- Adjust overhangs for manually designed primers while simulating Golden Gate Assembly
- Adjust the hybridization region for all PCR cloning simulations. When using automatic primer design, the hybridized region will be configured automatically, ensuring miscellaneous features are not transferred to the product when a 5’ primer extension by chance partially hybridizes to the template.
- Copy or export the properties and amino acid data for full protein sequences or selected regions
- Copy individual protein properties
Enhancements:
- Check for and prevent simulating Golden Gate assembly if two adjacent fragments are set to be used directly and abutting ends both lack 5' terminal phosphates
- Improved the appearance of many icons on low DPI displays
- Display the selected MW in the selection bar for protein alignments
- Improved the error message shown when attempting to ligate linear fragments that lack 5' terminal phosphates
- Updated the Golden Gate Assembly dialog to include easy access to recommended enzymes
- Improved the default sequence name when pasting FASTA encoded sequences into the New File dialog
- Modernized application, file, and various other icons
- Added links to NEBuilder® HiFi DNA Assembly and TOPO® Cloning tutorial videos.
- Show complementary bases of neighboring overhangs in Golden Gate cloning mini-overviews
- Zoom to fit by default when viewing RNA secondary structures
Fixes:
- Fixed an issue where quality of sequences aligned to a reference was sometimes not shown when requested
- Improved the appearance of tab controls on macOS
- Improved the appearance of slider controls on macOS and fixed an issue that prevented vertically scaling peaks for traces aligned to a reference sequence
- Improved overall stability
- Fixed an issue that prevented simulating Golden Gate cloning when a golden gate site blocked by methylation was present within the insert or in the assembled product
- Corrected an issue where when exporting translated features that use a custom genetic code (e.g. Amber) to GenBank the /transl_table qualifier should be omitted and this information is encoded using a /note qualifier instead
- Fixed issues with importing multi-sequence GenBank files that contain empty sequences (name only)
- Fixed an issue where protein sequences were sometimes imported from NCBI as DNA or RNA when using the import extra features option
- Fixed issues with selecting the codons just upstream and downstream of the site of ribosomal slippage
- Fixed an issue that sometimes prevented aligning bases adjacent to internal mismatching regions
- Ensure "Show History" is always a blue clickable link in textual representation of History view
- Corrected the default methylation for newly created sequences to match the default strain specified in Preferences
- Disabled the 3-Letter Amino Acids action in the sequence view context menu when using compact mode
- Fixed a crash when detecting common features
- Fixed an issue where quality data was sometimes not shown for all sequences aligned to a reference
- Fixed an issue with stripping out some formatting when copy and pasting from web pages into the description panel or other rich text controls
- Corrected an issue where feature colors were not accurately shown when using dark mode
- Addressed a number of issues with creating features in protein sequences
- Only allow a single copy of the alignment dialogs to be shown at a time
- Fixed the horizontal alignment of the "Show as Uninterrupted Circle" button in the side toolbar
- Do not show a cursor when clicking and holding on a codon in Sequence view
- Avoid showing duplicate copies of open documents from the Window menu
- Do not show a translation window when starting SnapGene on a computer configured to use a non-English locale such as German but one that the application is not translated into
- Removed duplicate "site" feature type from the feature type cascading menu
- Improve area printed when printing RNA structures
- Fixed an issue where printed RNA structures were sometimes blurry
- Fixed an issue where an I-beam cursor was shown when mousing over Sequence view in contexts where arbitrary selections cannot be made such as the Silent Mutagenesis dialog
- Fixed an issue that prevented detecting the preferred language on macOS
- Fixed an issue where the wrong number of binding sites was sometimes listed in cloning dilaogs when simplified binding sites were shown
- Fixed an issue where primers designed automatically and those specified manually were not always shown annealing to the template in the same manner
- Fixed various issues when using multiple screens.
- Various textual corrections
- Fixed a stability issue when attempting to use a linear fragment directly when simulating Overlap Extension PCR
- Fixed an issue where the application would freeze when attempting to generate primers for Golden Gate cloning if the fragments already contained one or more sites for the Golden Gate enzyme.
- Fixed issues where windows were not always shown on the logical display when using multiple displays on macOS.
- Improved opening older Geneious files.
- Fixed an issue where alternate transcripts (isoforms) were incorectly listed already in and thus not imported directly when importing features from another file.
- Fixed an issue that could prevent batch importing features
- Fixed an issue where when importing an enzyme list unknown entries were imported as AanI.
- Fixed an issue where using the Choose Primers command would fail to design primers for linear vectors or fragments if you did not first make a selection. SnapGene now correctly designers primers to use the entire linear sequence.
- Fixed a stability issue when changing the number of fragments to 1.
- Fixed an issue where if the first or last fragment in Overalap Extension PCR was not amplified by PCR the forward and/or reverse primers for amplifying the assembled linear fragment were not configured automatically when using the Choose Primers command.
- Fixed ordering from Vector Builder
- Fixed the link for more information about fonts and printing on Windows
- Improved rendering buttons and images when dragging windows between low and high resolution displays
- Fixed various memory leaks
- Fixed an issue where primers binding sites with melting temperatures less than 30 C were not identified
- Improved the behavior of the Secondary Structure zoom control
- Improved decoding feature types from Vector NTI databases
- Removed the MAFFT penalty shift setting since it was problematic
- Updated links to the user guide and user guide articles













